Machine Learning Module in OSCR
OSCR Machine Learning in Python
Linear Regression Module
© Kaixin Wang, Fall 2019
A PDF version of the tutorial can be accessed here
Module/Package import
import numpy as np # numpy module for linear algebra
import pandas as pd # pandas module for data manipulation
import matplotlib.pyplot as plt # module for plotting
import seaborn as sns # another module for plotting
import warnings # to handle warning messages
warnings.filterwarnings('ignore')
from sklearn.linear_model import LinearRegression # package for linear model
import statsmodels.api as sm # another package for linear model
import statsmodels.formula.api as smf
import scipy as sp
from sklearn.model_selection import train_test_split # split data into training and testing sets
Dataset import
The dataset that we will be using is the meuse dataset.
As described by the author of the data: “This data set gives locations and topsoil heavy metal concentrations, along with a number of soil and landscape variables at the observation locations, collected in a flood plain of the river Meuse, near the village of Stein (NL). Heavy metal concentrations are from composite samples of an area of approximately 15 m $\times$ 15 m.”
soil = pd.read_csv("soil.csv") # data import
soil.head() # check if read in correctly
| x | y | cadmium | copper | lead | zinc | elev | dist | om | ffreq | soil | lime | landuse | dist.m | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 181072 | 333611 | 11.7 | 85 | 299 | 1022 | 7.909 | 0.001358 | 13.6 | 1 | 1 | 1 | Ah | 50 |
| 1 | 181025 | 333558 | 8.6 | 81 | 277 | 1141 | 6.983 | 0.012224 | 14.0 | 1 | 1 | 1 | Ah | 30 |
| 2 | 181165 | 333537 | 6.5 | 68 | 199 | 640 | 7.800 | 0.103029 | 13.0 | 1 | 1 | 1 | Ah | 150 |
| 3 | 181298 | 333484 | 2.6 | 81 | 116 | 257 | 7.655 | 0.190094 | 8.0 | 1 | 2 | 0 | Ga | 270 |
| 4 | 181307 | 333330 | 2.8 | 48 | 117 | 269 | 7.480 | 0.277090 | 8.7 | 1 | 2 | 0 | Ah | 380 |
soil.shape # rows x columns
(155, 14)
print(soil.describe())
x y cadmium copper lead \
count 155.000000 155.000000 155.000000 155.000000 155.000000
mean 180004.600000 331634.935484 3.245806 40.316129 153.361290
std 746.039775 1047.746801 3.523746 23.680436 111.320054
min 178605.000000 329714.000000 0.200000 14.000000 37.000000
25% 179371.000000 330762.000000 0.800000 23.000000 72.500000
50% 179991.000000 331633.000000 2.100000 31.000000 123.000000
75% 180629.500000 332463.000000 3.850000 49.500000 207.000000
max 181390.000000 333611.000000 18.100000 128.000000 654.000000
zinc elev dist om ffreq \
count 155.000000 155.000000 155.000000 153.000000 155.000000
mean 469.716129 8.165394 0.240017 7.478431 1.606452
std 367.073788 1.058657 0.197702 3.432966 0.734111
min 113.000000 5.180000 0.000000 1.000000 1.000000
25% 198.000000 7.546000 0.075687 5.300000 1.000000
50% 326.000000 8.180000 0.211843 6.900000 1.000000
75% 674.500000 8.955000 0.364067 9.000000 2.000000
max 1839.000000 10.520000 0.880389 17.000000 3.000000
soil lime dist.m
count 155.000000 155.000000 155.000000
mean 1.451613 0.283871 290.322581
std 0.636483 0.452336 226.799927
min 1.000000 0.000000 10.000000
25% 1.000000 0.000000 80.000000
50% 1.000000 0.000000 270.000000
75% 2.000000 1.000000 450.000000
max 3.000000 1.000000 1000.000000
index = pd.isnull(soil).any(axis = 1)
soil = soil[-index]
soil = soil.reset_index(drop = True)
soil.shape
(152, 14)
n = soil.shape[1]
Exploratory Data Analysis (EDA)
Let suppose that the variable we are intereted in is the variable lead.
1. Correlation Heatmap
plt.figure(figsize = (10, 8))
sns.heatmap(soil.corr(), annot = True, square = True, linewidths = 0.1)
plt.ylim(n-1, 0)
plt.xlim(0, n-1)
plt.title("Pearson Correlation Heatmap between variables")
plt.show()
soil.corr()["lead"].sort_values()
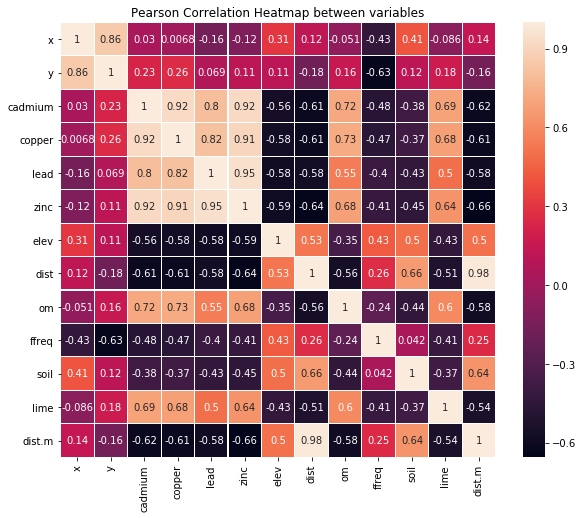
elev -0.584323
dist.m -0.584204
dist -0.576577
soil -0.430423
ffreq -0.399238
x -0.158104
y 0.069192
lime 0.501632
om 0.547836
cadmium 0.800898
copper 0.817000
zinc 0.954303
lead 1.000000
Name: lead, dtype: float64
plt.figure(figsize = (10, 8))
corr = soil.corr()
mask = np.zeros_like(corr)
mask[np.triu_indices_from(mask)] = True
with sns.axes_style("white"):
sns.heatmap(corr, mask = mask, linewidths = 0.1, vmax = .3, square = True)
plt.ylim(n-1, 0)
plt.xlim(0, n-1)
plt.title("correlation heatmap without symmetric information")
plt.show()
fig, axs = plt.subplots(figsize = (8, 12))
plt.subplots_adjust(left=0, bottom=0, right=1, top=1, wspace=0, hspace=0.2)
plt.subplot(2, 1, 1)
# correlation heatmap without annotation
sns.heatmap(soil.corr(), linewidths = 0.1, square = True, cmap = "YlGnBu")
plt.ylim(n-1, 0)
plt.xlim(0, n-1)
plt.title("correlation heatmap without annotation")
plt.subplot(2, 1, 2)
# correlation heatmap with annotation
sns.heatmap(soil.corr(), linewidths = 0.1, square = True, annot = True, cmap = "YlGnBu")
plt.ylim(n-1, 0)
plt.xlim(0, n-1)
plt.title("correlation heatmap with annotation")
plt.show()
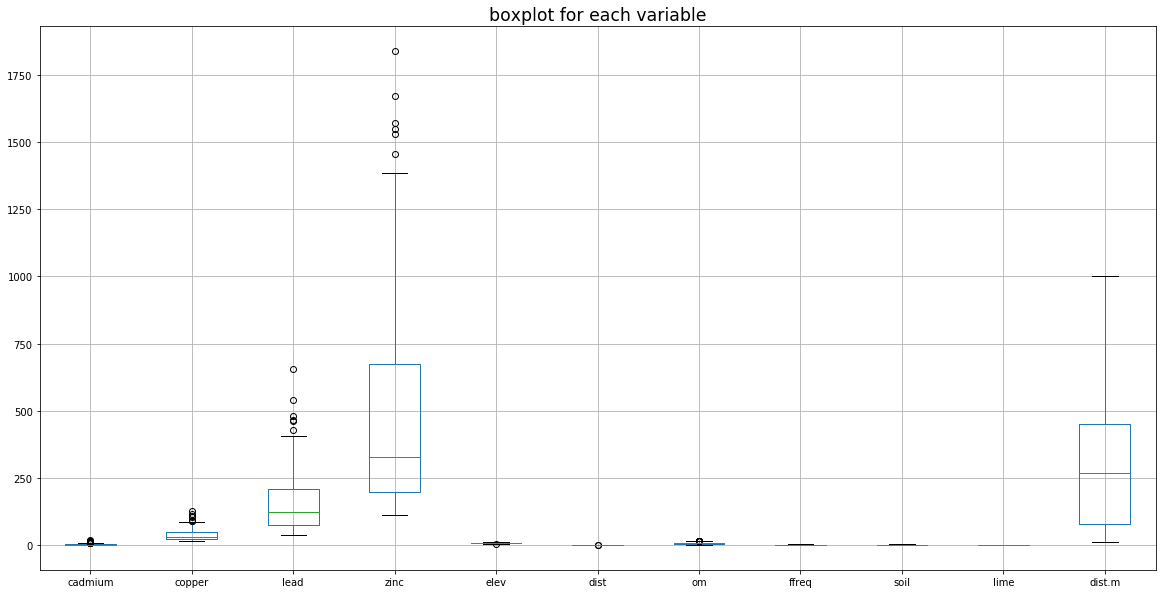
2. Boxplots
plt.figure(figsize = (20, 10))
soil.iloc[:, 2:].boxplot()
# soil.iloc[:, 2:].plot(kind = "box") # 2nd method
plt.title("boxplot for each variable", fontsize = "xx-large")
plt.show()
fig, axs = plt.subplots(2, int(n/2), figsize = (20, 10))
colors = ['b', 'g', 'r', 'c', 'm', 'y', 'pink'] # to set color
for i, var in enumerate(soil.columns.values):
if var == "landuse":
axs[1, i-int(n/2)].set_xlabel(var, fontsize = 'large')
continue
if i < int(n/2):
axs[0, i].boxplot(soil[var])
axs[0, i].set_xlabel(var, fontsize = 'large')
axs[0, i].scatter(x = np.tile(1, soil.shape[0]), y = soil[var], color = colors[i])
else:
axs[1, i-int(n/2)].boxplot(soil[var])
axs[1, i-int(n/2)].set_xlabel(var, fontsize = 'large')
axs[1, i-int(n/2)].scatter(x = np.tile(1, soil.shape[0]), y = soil[var],
color = colors[i-int(n/2)])
plt.suptitle("boxplot of variables", fontsize = 'xx-large')
plt.show()
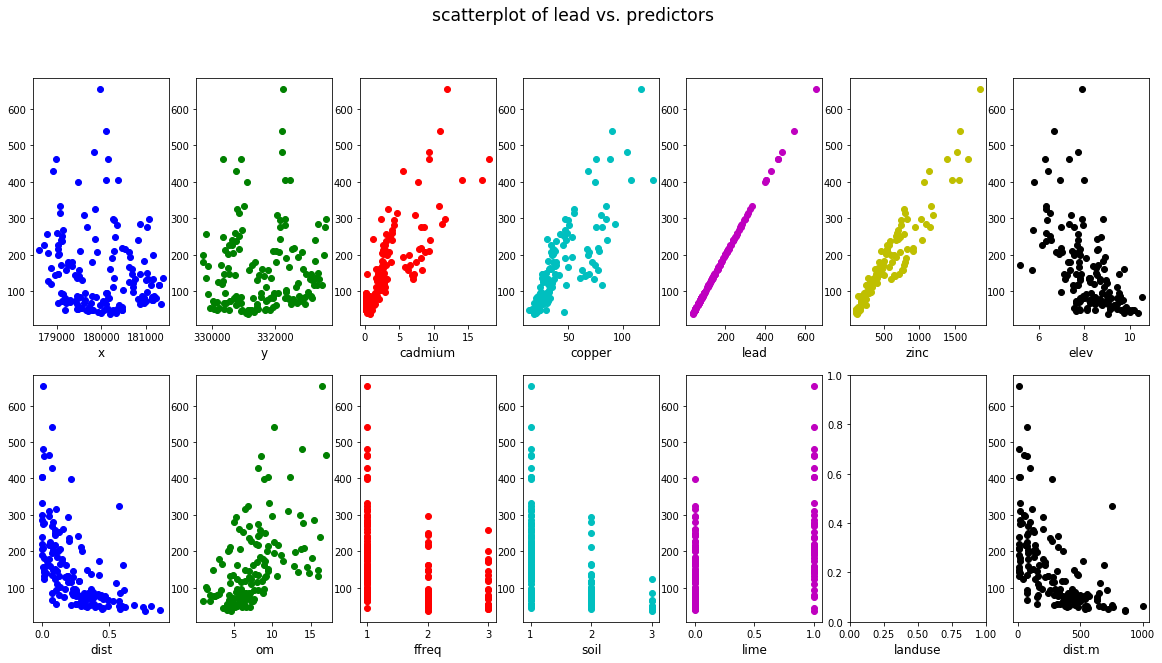
3. scatterplots
fig, axs = plt.subplots(2, int(n/2), figsize = (20, 10))
colors = ['b', 'g', 'r', 'c', 'm', 'y', 'k'] # to set color
for i, var in enumerate(soil.columns.values):
if var == "landuse":
axs[1, i-int(n/2)].set_xlabel(var, fontsize = 'large')
continue
if i < int(n/2):
axs[0, i].scatter(soil[var], soil["lead"], color = colors[i])
axs[0, i].set_xlabel(var, fontsize = 'large')
else:
axs[1, i-int(n/2)].scatter(soil[var], soil["lead"], color = colors[i-int(n/2)])
axs[1, i-int(n/2)].set_xlabel(var, fontsize = 'large')
plt.suptitle("scatterplot of lead vs. predictors", fontsize = 'xx-large')
plt.show()
4. histograms and density plots
fig, axs = plt.subplots(2, int(n/2), figsize = (20, 10))
colors = ['b', 'g', 'r', 'c', 'm', 'y', 'pink'] # to set color
for i, var in enumerate(soil.columns.values):
if var == "landuse":
axs[1, i-int(n/2)].set_xlabel(var, fontsize = 'large')
continue
if i < int(n/2):
sns.distplot(soil[var], color = colors[i], ax = axs[0, i])
else:
sns.distplot(soil[var], color = colors[i-int(n/2)], ax = axs[1, i-int(n/2)])
plt.suptitle("histogram and density plot of each variable", fontsize = 'xx-large')
plt.show()
Variable Selection and Modeling
Based on the scatterplot of lead vs. predictors:
cadmium,copper,zincandomseem to have strong positive correlation with leadelev,distanddist.mseem to have strong negative correlation with lead- categorical variable
limeseems to be a good indicator predictor variable
Therefore, we now build a linear model using 6 predictors: cadmium, copper, zinc, elev, dist and lime.
# variables = ["cadmium", "copper", "zinc", "elev", "dist", "dist.m", "lime"]
variables = ["cadmium", "copper", "zinc", "elev", "dist", "lime"]
x = soil[variables]
y = soil["lead"]
Modeling - building a linear model
split the dataset into training and testing set
X_train, X_test, y_train, y_test = train_test_split(x, y, test_size = 0.33, random_state = 42)
build a linear model using sklearn package
model = LinearRegression().fit(X_train, y_train)
# R^2 on training set
R2_train = model.score(X_train, y_train)
R2_train
0.9483947080189404
# R^2 on testing set
R2_test = model.score(X_test, y_test)
R2_test
0.964901551899052
# value of intercepts
for var, coef in zip(variables, model.coef_):
print(var, coef)
cadmium -13.391990002100986
copper 0.05365863276389149
zinc 0.42346584485869393
elev -3.2531127698493183
dist 14.509965024252311
lime -23.514058405476963
build a linear model using statsmodel package
df_train = pd.concat([X_train, y_train], axis = 1) # build a dataframe for training set
fullModel = smf.ols("lead ~ cadmium + copper + zinc + elev + C(lime)", data = df_train)
fullModel = fullModel.fit()
print(fullModel.summary())
OLS Regression Results
==============================================================================
Dep. Variable: lead R-squared: 0.948
Model: OLS Adj. R-squared: 0.945
Method: Least Squares F-statistic: 346.6
Date: Tue, 12 Nov 2019 Prob (F-statistic): 2.35e-59
Time: 08:17:24 Log-Likelihood: -467.76
No. Observations: 101 AIC: 947.5
Df Residuals: 95 BIC: 963.2
Df Model: 5
Covariance Type: nonrobust
================================================================================
coef std err t P>|t| [0.025 0.975]
--------------------------------------------------------------------------------
Intercept 23.8739 27.200 0.878 0.382 -30.126 77.874
C(lime)[T.1] -24.7610 7.775 -3.185 0.002 -40.196 -9.326
cadmium -13.2322 2.240 -5.908 0.000 -17.679 -8.785
copper 0.0522 0.278 0.187 0.852 -0.500 0.605
zinc 0.4188 0.020 20.756 0.000 0.379 0.459
elev -2.4719 2.895 -0.854 0.395 -8.220 3.276
==============================================================================
Omnibus: 2.077 Durbin-Watson: 1.947
Prob(Omnibus): 0.354 Jarque-Bera (JB): 1.494
Skew: -0.246 Prob(JB): 0.474
Kurtosis: 3.336 Cond. No. 6.42e+03
==============================================================================
Warnings:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
[2] The condition number is large, 6.42e+03. This might indicate that there are
strong multicollinearity or other numerical problems.
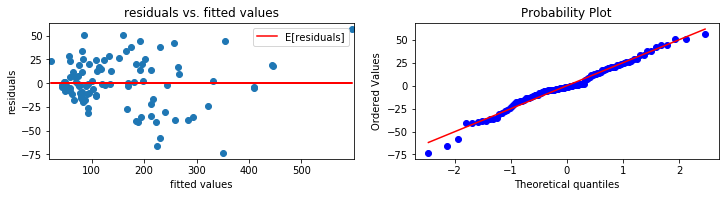
diagnostics plots
# diagnosticsPlots function definition
def diagnosticsPlots(predictions, truevalues):
'''
This function will plot the following two diagonistics plots:
1. residuals vs. fitted values plot (plot on the left)
2. qqplot of the residuals (plot on the right)
parameters required:
- predictions: predicted values using the linear regression model
- truevalues: true values of the response variable
required modules/packages:
- matplotlib.pyplot (plt)
- scipy (sp)
Author: Kaixin Wang
Created: October 2019
'''
residuals = truevalues - predictions
fig, axs = plt.subplots(figsize = (12, 2.5))
plt.subplot(1, 2, 1)
# residuals vs. fitted values
plt.scatter(predictions, residuals)
plt.plot(predictions, np.tile(np.mean(residuals), residuals.shape[0]), "r-")
plt.xlim([np.min(predictions) - 2.5, np.max(predictions) + 2.5])
plt.title("residuals vs. fitted values")
plt.ylabel("residuals")
plt.xlabel("fitted values")
plt.legend(["E[residuals]"])
print("Average of residuals: ", np.mean(residuals))
# qqplot
plot = plt.subplot(1, 2, 2)
sp.stats.probplot(residuals, plot = plot, fit = True)
plt.show()
pred_val = fullModel.fittedvalues.copy()
true_val = y_train.copy()
diagnosticsPlots(pred_val, true_val)
Average of residuals: 2.583420825998257e-12
training and testing Root-Mean-Square Error (RMSE)
The root-mean-square error of a prediction is calcuated as the following:
where $y_i$ is the true value of the response, $\hat {y_i}$ is the fitted value of the response, n is the size of the input
def RMSETable(train_y, train_fitted, train_size, train_R2, test_y, test_fitted, test_size, test_R2):
'''
This function creates a function that returns a dataframe in the following format:
-------------------------------------------------
RMSE R-squared size
training set train_RMSE train_R2 n_training
testing set test_RMSE tesint_R2 n_testing
-------------------------------------------------
parameters required:
- train_y: true values of the response in the training set
- train_fitted: fitted values of the response in the training set
- train_size: size of the training set
- train_R2: R-squared on the training set
- test_y: true values of the response in the testing set
- test_fitted: fitted values of the response in the testing set
- test_size: size of the testing size
- test_R2: R-squared on the testing set
Author: Kaixin Wang
Created: October 2019
'''
train_RMSE = np.sqrt(sum(np.power(train_y - train_fitted, 2))) / train_size
test_RMSE = np.sqrt(sum(np.power(test_y - test_fitted, 2))) / test_size
train = [train_RMSE, train_size, train_R2]
test = [test_RMSE, test_size, test_R2]
return pd.DataFrame([train, test], index = ["training set", "testing set"], columns = ["RMSE", "R-squared", "size"])
table1 = RMSETable(y_train.values, model.predict(X_train), R2_train, len(y_train),
y_test.values, model.predict(X_test), R2_test, len(y_test))
print(table1)
RMSE R-squared size
training set 262.286912 0.948395 101
testing set 159.247070 0.964902 51
Model selection
Since we are using 5 predictor variables for 152 entries, we can consider using fewer predictors to improve the $R_{adj}^2$, where is a version of $R^2$ that penalizes model complexity.
Selecting fewer predictor will also help with preventing overfitting.
Therefore, observing that zinc, copper and cadmium have relatively stronger correlation with lead, we now build a model using only these three predictors.
using sklearn linear regression module
variables = ["cadmium", "lime", "zinc"]
x = soil[variables]
y = soil["lead"]
X_train, X_test, y_train, y_test = train_test_split(x, y, test_size = 0.33, random_state = 42)
model2 = LinearRegression().fit(X_train, y_train)
# R^2 on training set
R2_train = model2.score(X_train, y_train)
print("Training set R-squared", R2_train)
# R^2 on testing set
R2_test = model2.score(X_test, y_test)
print("Testing set R-squared", R2_test)
Training set R-squared 0.9475953381623881
Testing set R-squared 0.9609072316593821
# value of intercepts
for var, coef in zip(variables, model2.coef_):
print(var, coef)
cadmium -13.093360663968943
lime -23.487105420923648
zinc 0.4236873112817108
using statsmodel ordinary linear regrssion module
df_train = pd.concat([X_train, y_train], axis = 1) # build a dataframe for training set
reducedModel = smf.ols("lead ~ cadmium + C(lime) + zinc", data = df_train)
reducedModel = reducedModel.fit()
print(reducedModel.summary())
OLS Regression Results
==============================================================================
Dep. Variable: lead R-squared: 0.948
Model: OLS Adj. R-squared: 0.946
Method: Least Squares F-statistic: 584.7
Date: Tue, 12 Nov 2019 Prob (F-statistic): 5.98e-62
Time: 08:17:24 Log-Likelihood: -468.19
No. Observations: 101 AIC: 944.4
Df Residuals: 97 BIC: 954.8
Df Model: 3
Covariance Type: nonrobust
================================================================================
coef std err t P>|t| [0.025 0.975]
--------------------------------------------------------------------------------
Intercept 2.6976 4.614 0.585 0.560 -6.461 11.856
C(lime)[T.1] -23.4871 7.569 -3.103 0.003 -38.509 -8.465
cadmium -13.0934 1.986 -6.593 0.000 -17.035 -9.152
zinc 0.4237 0.019 22.540 0.000 0.386 0.461
==============================================================================
Omnibus: 0.749 Durbin-Watson: 1.968
Prob(Omnibus): 0.688 Jarque-Bera (JB): 0.395
Skew: -0.131 Prob(JB): 0.821
Kurtosis: 3.160 Cond. No. 1.79e+03
==============================================================================
Warnings:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
[2] The condition number is large, 1.79e+03. This might indicate that there are
strong multicollinearity or other numerical problems.
diagnostics plots
pred_val = reducedModel.fittedvalues.copy()
true_val = y_train.copy()
diagnosticsPlots(pred_val, true_val)
Average of residuals: 3.392313932510102e-13
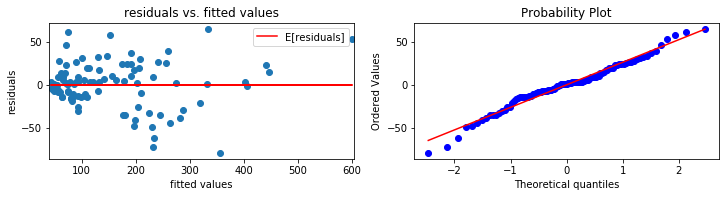
table2 = RMSETable(y_train.values, model2.predict(X_train), R2_train, len(y_train),
y_test.values, model2.predict(X_test), R2_test, len(y_test))
print(table2)
RMSE R-squared size
training set 264.533494 0.947595 101
testing set 168.763005 0.960907 51
model evaluation and comparison
- model with 5 predictors (full model)
print(table1)
RMSE R-squared size
training set 262.286912 0.948395 101
testing set 159.247070 0.964902 51
- model with 3 predictors (reduced model)
print(table2)
RMSE R-squared size
training set 264.533494 0.947595 101
testing set 168.763005 0.960907 51
table = sm.stats.anova_lm(reducedModel, fullModel)
table
| df_resid | ssr | df_diff | ss_diff | F | Pr(>F) | |
|---|---|---|---|---|---|---|
| 0 | 97.0 | 62835.802512 | 0.0 | NaN | NaN | NaN |
| 1 | 95.0 | 62309.421080 | 2.0 | 526.381432 | 0.401273 | 0.670596 |
Comparing the tables obtained above:
- The full model with 6 predictors has lower test RMSE and higher $R^2$ value than the reduced model with 3 predictors
- However, a higher $R^2$ doesn’t mean that we favor the full model, since $R^2$ will always increase as the number of predictors used increases. Instead, we will take a look at $R^2_{adj}$, the adjusted $R^2$, which is calculated as
where $n$ is the sample size and $p$ is the number of predictors used in the model.
Since $R_{adj}^2$ of the full model is 0.945, while the $R_{adj}^2$ of the reduced model is 0.946, which is an improvement, we observe that the reduced model has a lower model complexity while maintaining a good prediction performance.
Summary of the module
By building two linear regression models using 6 and 3 predictors respectively, we observe that predictor variables cadmium, copper and zinc have very strong positive correlation with the response variable lead. In addition, we also observed that by adding three more predictors, dist, elev and lime, the $R^2_{adj}$ didn’t get much improvement comparing to the model with only 3 predictors.
Therefore, the final model that we decided to adopt is the reduced model with 3 predictors:
Reference:
Dataset: meuse dataset
- P.A. Burrough, R.A. McDonnell, 1998. Principles of Geographical Information Systems. Oxford University Press.
- Stichting voor Bodemkartering (STIBOKA), 1970. Bodemkaart van Nederland : Blad 59 Peer, Blad 60 West en 60 Oost Sittard: schaal 1 : 50 000. Wageningen, STIBOKA.
To access this dataset in R:
install.packages("sp")
library(sp)
data(meuse)
© Kaixin Wang, updated October 2019
